TNA also enables the analysis of transition networks constructed from
grouped sequence data. In this example, we first fit a mixed Markov
model to the engagement data using the seqHMM
package and build a grouped TNA model based on this model. First, we
load the packages we will use for this example.
library("tna")
library("tibble")
library("dplyr")
library("gt")
library("seqHMM")
data("engagement", package = "tna")We simulate transition probabilities to initialize the model.
set.seed(265)
tna_model <- tna(engagement)
n_var <- length(tna_model$labels)
n_clusters <- 3
trans_probs <- simulate_transition_probs(n_var, n_clusters)
init_probs <- list(
c(0.70, 0.20, 0.10),
c(0.15, 0.70, 0.15),
c(0.10, 0.20, 0.70)
)Next, we building and fit the model (this step takes some time to
compute, the final model object is also available in the
tna package as engagement_mmm).
mmm <- build_mmm(
engagement,
transition_probs = trans_probs,
initial_probs = init_probs
)
fit_mmm <- fit_model(
modelTrans,
global_step = TRUE,
control_global = list(algorithm = "NLOPT_GD_STOGO_RAND"),
local_step = TRUE,
threads = 60,
control_em = list(restart = list(times = 100, n_optimum = 101))
)Now, we create a new model using the cluster information from the
model. Alternatively, if sequence data is provided to
group_model(), the group assignments can be provided with
the group argument.
tna_model_clus <- group_model(fit_mmm$model)We can summarize the cluster-specific models
summary(tna_model_clus) |>
gt() |>
fmt_number(decimals = 2)| metric | Cluster 1 | Cluster 2 | Cluster 3 |
|---|---|---|---|
| Node Count | 3.00 | 3.00 | 3.00 |
| Edge Count | 9.00 | 8.00 | 8.00 |
| Network Density | 1.00 | 1.00 | 1.00 |
| Mean Distance | 0.11 | 0.24 | 0.30 |
| Mean Out-Strength | 1.00 | 1.00 | 1.00 |
| SD Out-Strength | 0.21 | 0.35 | 0.47 |
| Mean In-Strength | 1.00 | 1.00 | 1.00 |
| SD In-Strength | 0.00 | 0.00 | 0.00 |
| Mean Out-Degree | 3.00 | 2.67 | 2.67 |
| SD Out-Degree | 0.00 | 0.58 | 0.58 |
| Centralization (Out-Degree) | 0.00 | 0.25 | 0.25 |
| Centralization (In-Degree) | 0.00 | 0.25 | 0.25 |
| Reciprocity | 1.00 | 0.80 | 0.80 |
and their initial probabilities
bind_rows(lapply(tna_model_clus, \(x) x$inits), .id = "Cluster") |>
gt() |>
fmt_percent()| Cluster 1 | Cluster 2 | Cluster 3 |
|---|---|---|
| 33.98% | 75.00% | 0.00% |
| 32.35% | 8.33% | 0.00% |
| 33.67% | 16.67% | 100.00% |
as well as transition probabilities.
transitions <- lapply(
tna_model_clus,
function(x) {
x$weights |>
data.frame() |>
rownames_to_column("From\\To") |>
gt() |>
tab_header(title = names(tna_model_clus)[1]) |>
fmt_percent()
}
)
transitions[[1]]| Cluster 1 | |||
| From\To | Active | Average | Disengaged |
|---|---|---|---|
| Active | 85.99% | 8.92% | 5.09% |
| Average | 31.21% | 54.21% | 14.58% |
| Disengaged | 4.79% | 16.18% | 79.03% |
transitions[[2]]| Cluster 1 | |||
| From\To | Active | Average | Disengaged |
|---|---|---|---|
| Active | 84.09% | 15.91% | 0.00% |
| Average | 9.26% | 62.96% | 27.78% |
| Disengaged | 15.56% | 51.11% | 33.33% |
transitions[[3]]| Cluster 1 | |||
| From\To | Active | Average | Disengaged |
|---|---|---|---|
| Active | 58.33% | 12.50% | 29.17% |
| Average | 15.28% | 81.94% | 2.78% |
| Disengaged | 0.00% | 60.00% | 40.00% |
We can also plot the cluster-specific transitions
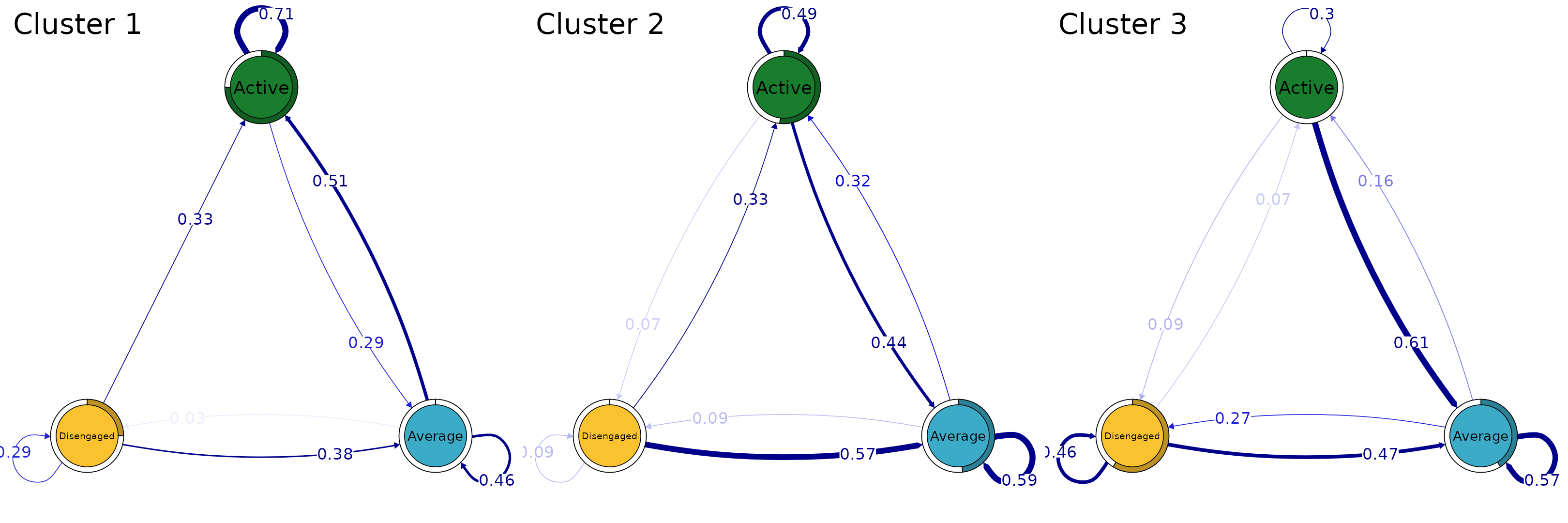
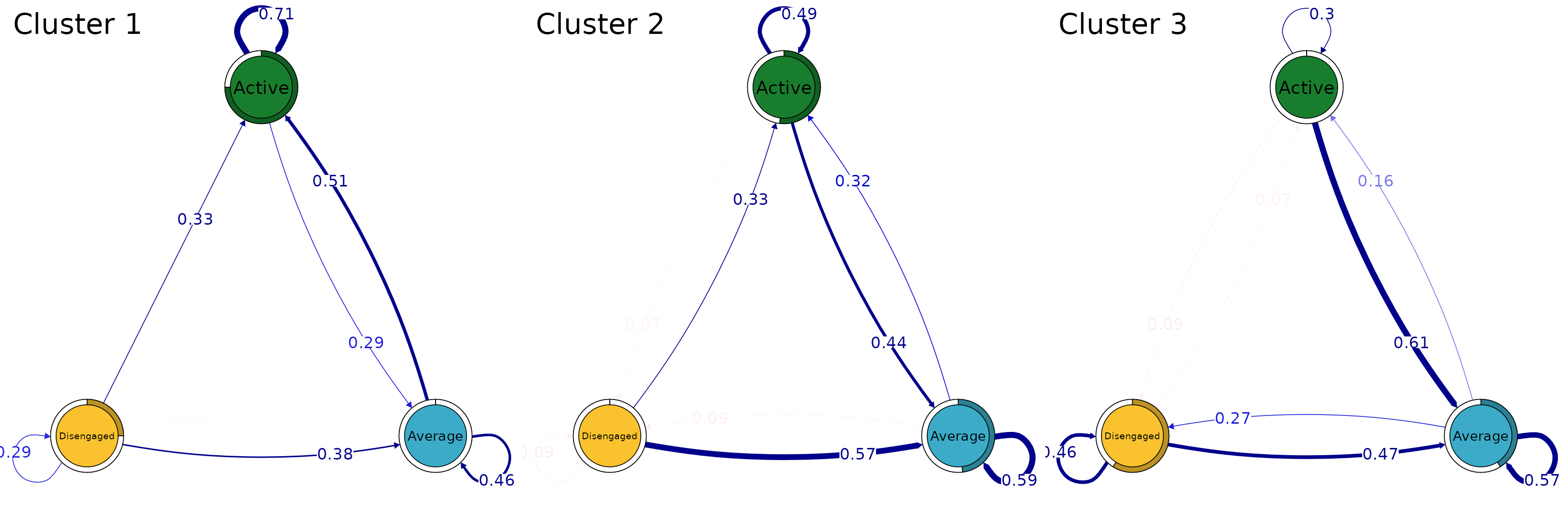
Just like ordinary TNA models, we can prune the rare transitions
pruned_clus <- prune(tna_model_clus, threshold = 0.1)and plot the cluster transitions after pruning
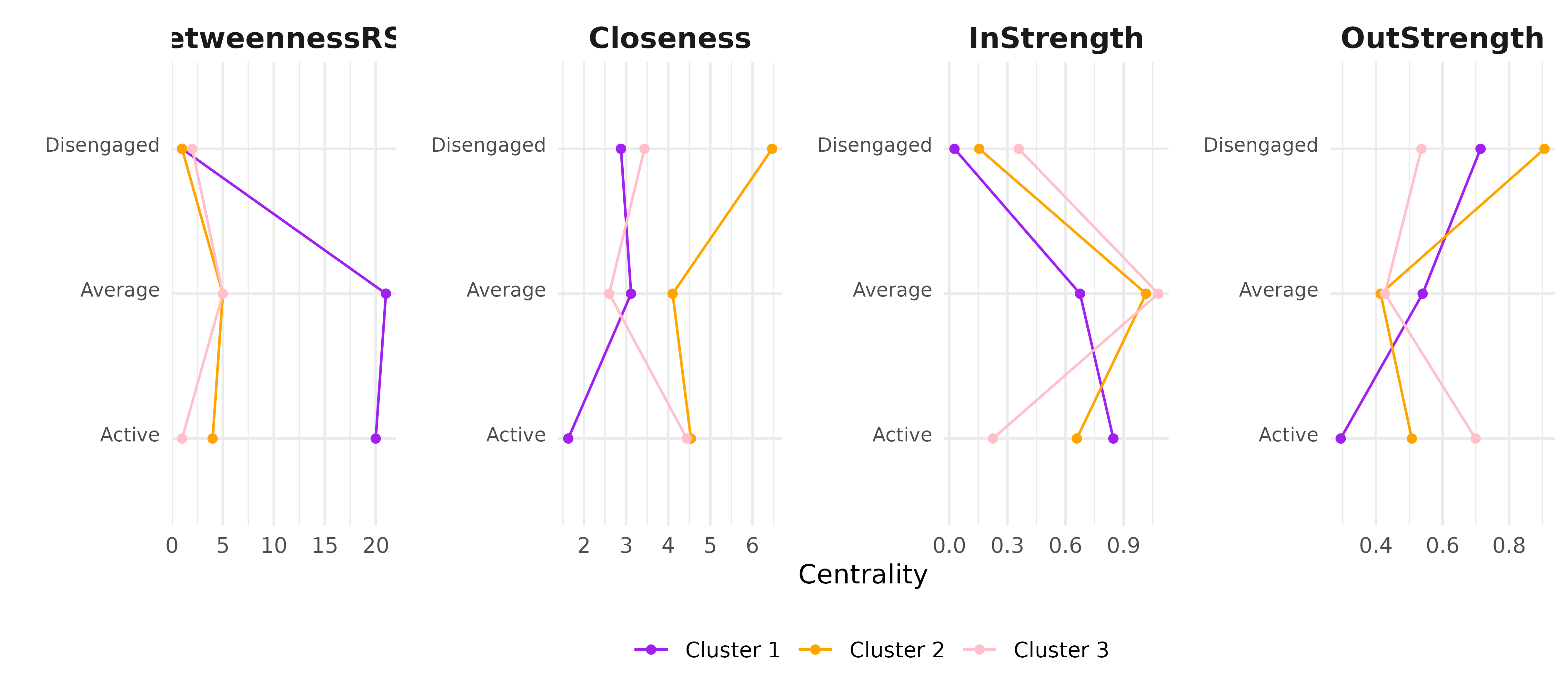
Centrality measures can also be computed for each cluster directly.
centrality_measures <- c(
"BetweennessRSP",
"Closeness",
"InStrength",
"OutStrength"
)
centralities_per_cluster <- centralities(
tna_model_clus,
measures = centrality_measures
)
plot(
centralities_per_cluster, ncol = 4,
colors = c("purple", "orange", "pink")
)