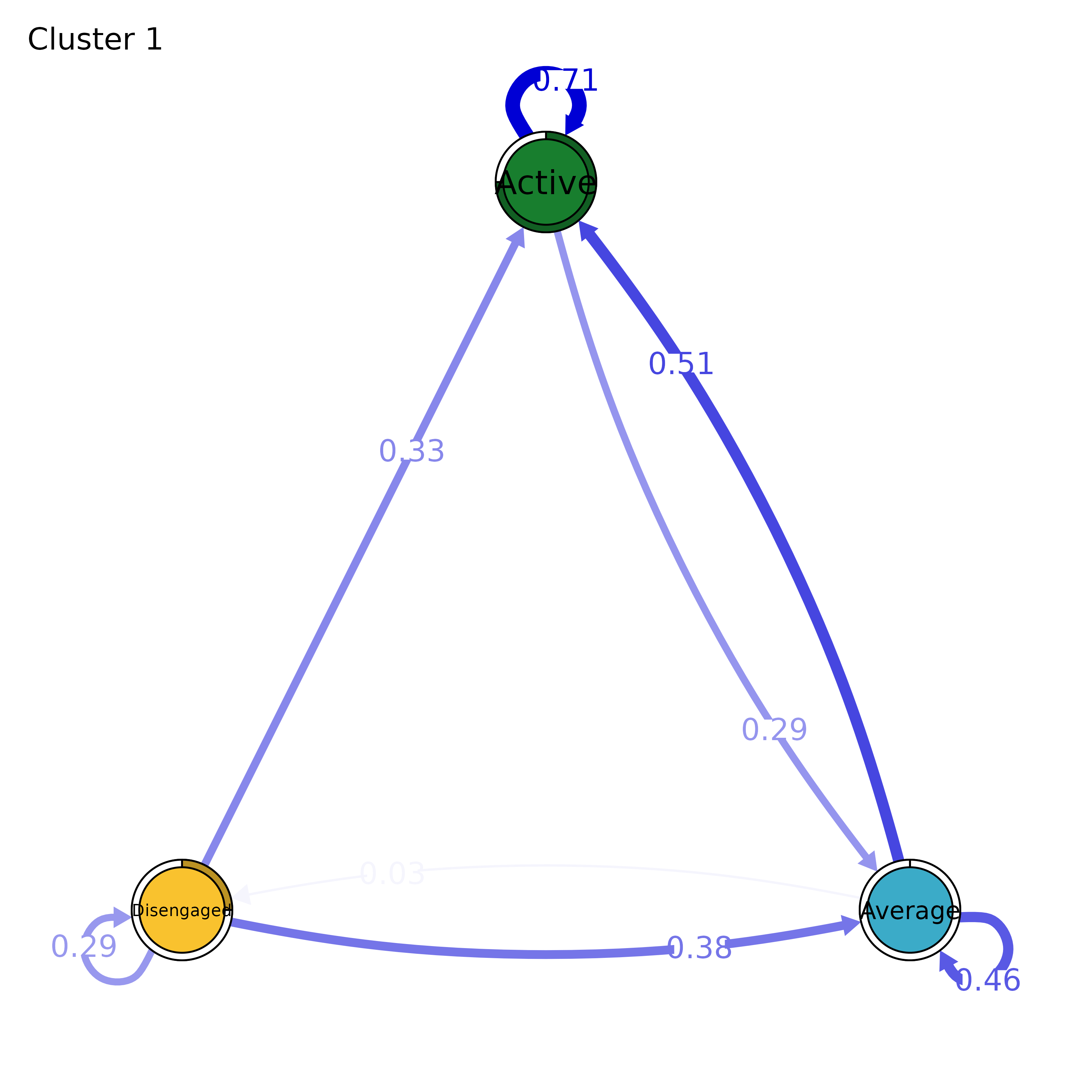
Plots a transition network of each cluster using qgraph.
Usage
# S3 method for class 'group_tna'
plot(x, title, which, ...)Arguments
- x
A
group_modelobject.- title
A title for each plot. It can be a single string (the same one will be used for all plots) or a list (one per group)
- which
An optional
integervector of groups to plot. By default, all groups are plotted.- ...
Arguments passed on to
plot.tnalabelsSee
qgraph::qgraph().colorsSee
qgraph::qgraph().pieSee
qgraph::qgraph().cutEdge color and width emphasis cutoff value. The default is the median of the edge weights. See
qgraph::qgraph()for details.show_prunedA
logicalvalue indicating if pruned edges removed byprune()should be shown in the plot. The default isTRUE, and the edges are drawn as dashed with a different color to distinguish them.pruned_edge_colorA
characterstring for the color to use for pruned edges whenshow_pruned = TRUE. The default is"pink".edge.colorSee
qgraph::qgraph().edge.labelsSee
qgraph::qgraph().edge.label.positionSee
qgraph::qgraph().layoutOne of the following:
A
characterstring describing aqgraphlayout (e.g.,"circle") or the name of aigraphlayout function (e.g.,"layout_on_grid").A
matrixof node positions to use, with a row for each node andxandycolumns for the node positions.A layout function from
igraph.
layout_argsA
listof arguments to pass to theigraphlayout function whenlayoutis a function or a character string that specifies a function name.scale_nodesA
characterstring giving the name of a centrality measure to scale the node size by. Seecentralities()for valid names. If missing (the default), uses defaultqgraph::qgraph()scaling. Overridesvsizeprovided via....scaling_factorA
numericvalue specifying how strongly to scale the nodes whenscale_nodesis provided. Values between 0 and 1 will result in smaller differences and values larger than 1 will result in greater differences. The default is0.5.marSee
qgraph::qgraph().themeSee
qgraph::qgraph().
See also
Basic functions
build_model(),
hist.group_tna(),
hist.tna(),
plot.tna(),
plot_frequencies(),
plot_frequencies.group_tna(),
plot_mosaic(),
plot_mosaic.group_tna(),
plot_mosaic.tna_data(),
print.group_tna(),
print.summary.group_tna(),
print.summary.tna(),
print.tna(),
summary.group_tna(),
summary.tna(),
tna-package
Examples
model <- group_model(engagement_mmm)
plot(model, which = 1)