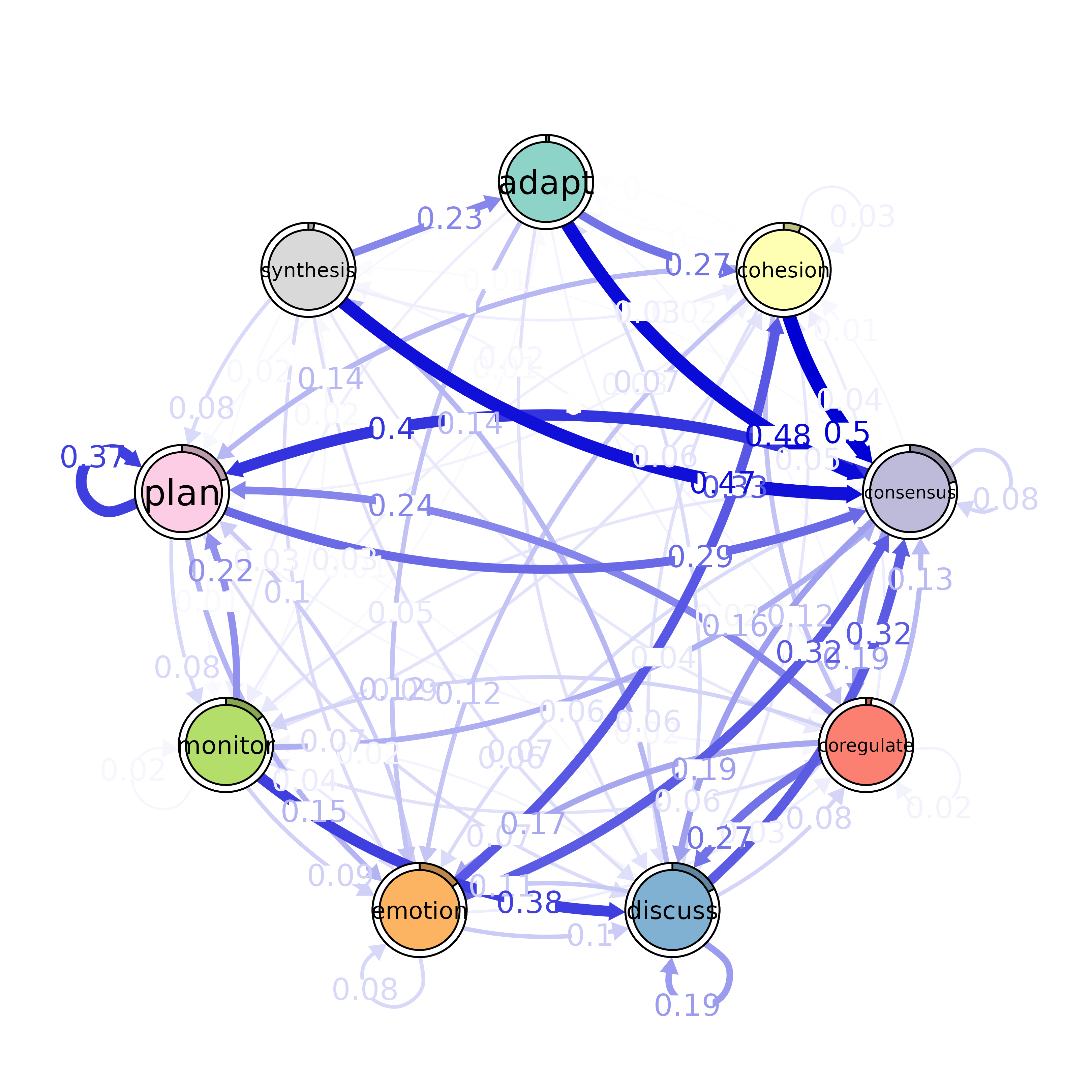
This function plots a transition network analysis (TNA) model using
the qgraph package. The nodes in the graph represent states, with node
sizes corresponding to initial state probabilities. Edge labels represent
the edge weights of the network.
Arguments
- x
A
tnaobject fromtna().- labels
See
qgraph::qgraph().- colors
See
qgraph::qgraph().- pie
See
qgraph::qgraph().- cut
Edge color and width emphasis cutoff value. The default is the median of the edge weights. See
qgraph::qgraph()for details.- show_pruned
A
logicalvalue indicating if pruned edges removed byprune()should be shown in the plot. The default isTRUE, and the edges are drawn as dashed with a different color to distinguish them.- pruned_edge_color
A
characterstring for the color to use for pruned edges whenshow_pruned = TRUE. The default is"pink".- edge.color
See
qgraph::qgraph().- edge.labels
See
qgraph::qgraph().- edge.label.position
See
qgraph::qgraph().- layout
One of the following:
A
characterstring describing aqgraphlayout (e.g.,"circle") or the name of aigraphlayout function (e.g.,"layout_on_grid").A
matrixof node positions to use, with a row for each node andxandycolumns for the node positions.A layout function from
igraph.
- layout_args
A
listof arguments to pass to theigraphlayout function whenlayoutis a function or a character string that specifies a function name.- scale_nodes
A
characterstring giving the name of a centrality measure to scale the node size by. Seecentralities()for valid names. If missing (the default), uses defaultqgraph::qgraph()scaling. Overridesvsizeprovided via....- scaling_factor
A
numericvalue specifying how strongly to scale the nodes whenscale_nodesis provided. Values between 0 and 1 will result in smaller differences and values larger than 1 will result in greater differences. The default is0.5.- mar
See
qgraph::qgraph().- theme
See
qgraph::qgraph().- ...
Additional arguments passed to
qgraph::qgraph().
See also
Basic functions
build_model(),
hist.group_tna(),
hist.tna(),
plot.group_tna(),
plot_frequencies(),
plot_frequencies.group_tna(),
plot_mosaic(),
plot_mosaic.group_tna(),
plot_mosaic.tna_data(),
print.group_tna(),
print.summary.group_tna(),
print.summary.tna(),
print.tna(),
summary.group_tna(),
summary.tna(),
tna-package